In September 2015, GARNet and OpenPlant organized a two-day workshop at the John Innes Centre that provided both background information and hands-on training for CRISPR-Cas technology. The report from that meeting is now online, co-authored by Dr Nicola Patron and Dr Colette Matthewman from OpenPlant along with GARNet colleagues.
Parry, G., Patron, N., Bastow, R., & Matthewman, C. (2016). Meeting report: GARNet/OpenPlant CRISPR-Cas workshop. Plant methods, 12(1), 1.
Full text
PDF (1.2MB)
The article is fully Open Access under a CC-BY 4.0 license so it’s reproduced below!
Meeting report: GARNet/OpenPlant CRISPR-Cas workshop
- Geraint Parry†Email author,
- Nicola Patron†,
- Ruth Bastow and
- Colette Matthewman
†Contributed equally
Plant Methods201612:6
DOI: 10.1186/s13007-016-0104-
© Parry et al. 2016
Received: 17 November 201
Accepted: 5 January 2016
Published: 27 January 2016
Abstract
Targeted genome engineering has been described as a “game-changing technology” for fields as diverse as human genetics and plant biotechnology. One technique used for precise gene editing utilises the CRISPR-Cas system and is an effective method for genetic engineering in a wide variety of plants. However, many researchers remain unaware of both the technical challenges that emerge when using this technique or of its potential benefits. Therefore in September 2015, GARNet and OpenPlant organized a two-day workshop at the John Innes Centre that provided both background information and hands-on training for this important technology.
Keywords
CRISPR Cas9 Gene Editing Genetic Engineering
Geraint Parry and Nicola Patron contributed equally to this manuscript
Background
Over the past few years, genome engineering (GE), the process of making targeted modifications to the genome, its contexts or its outputs, has been described as a “game-changing technology” for fields as diverse as human genetics and plant biotechnology. The ability to introduce specific changes to genomic loci adds a level of precision not previously available to molecular biologists working in multicellular eukaryotes. Despite overwhelming scientific opinion that Genetically Modified (GM) plants are safe and provide environmental and socioeconomic benefits, they remain broadly unpopular outside of the scientific community [1–3]. This has been blamed both on inaccurate media reporting and public concerns over the ownership of technologies that underpin food production [4–6]. Given these political and public opinions, plant scientists are particularly hopeful about the future use of GE technologies, which are likely to enable precise genetic changes to be made without the ongoing requirement for foreign DNA to be integrated the genome.
However, despite some countries ruling that plants with targeted mutations may not be regulated as GM, there is still much uncertainty [7, 8]. Even as the technologies behind GE are being optimized, the scientific community is engaging with stakeholders to highlight potential positive uses, including how it might be used to develop better crops. This is exemplified by a policy statement from the UK’s Biotechnology and Biological Sciences Research Council (BBSRC) on “New Techniques for Genetic Crop Improvement” that outlines positive uses for GE technologies [9].
The experimental protocols needed to implement these powerful techniques are yet to be embraced by many plant science laboratories. To address this issue GARNet [10] and OpenPlant [11] collaborated to organise a workshop to explain the background of Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/Cas technologies for GE in plants and to equip plant scientists with the skills required to implement Cas9-induced targeted mutagenesis. Over 140 researchers registered for the meeting, held at the John Innes Centre (UK), from as far-afield as Ireland and Poland, clearly demonstrating the appetite to apply these technologies to plant systems. The first day was open to all attendees and consisted of conventional ‘seminar-style’ presentations, while day two was a hands-on introduction for 30 researchers. This meeting was made possible by the kind support of Plant Methods.
Day one presentations
The meeting was opened by Dr. Jim Haseloff from The University of Cambridge who introduced synthetic biology in plant systems and Dr. Nicola Patron of The Sainsbury Laboratory, Norwich (TSL), the primary organiser, who provided a historical perspective on GE technologies. The specifics of these technologies are discussed in detail as part of this Plant Methodsthematic series. Keynote presentations were given by Professor Holger Puchta (Karlsruhe Institute of Technology) and Professor Bing Yang (Iowa State University) who each provided overviews and success stories from their own laboratories. These were followed by shorter talks from scientists at JIC, TSL and the University of Cambridge who are already working with CRISPR/Cas technologies.
Professor Puchta gave an inspiring talk that provided attendees with the history of his seminal work. He presented earlier work showing that induction of double strand breaks (DSBs) using site-specific endonucleases can enhance the freqeuncy of homologous recombination in plant cells through to his recent work using RNA-guided Cas9 nuclease to induce DSBs [12, 13]. He mentioned that the two most important molecular discoveries of his lifetime had been the Polymerase Chain Reaction (PCR) and GE technologies, the latter he described as having “hit him like a tsunami”. It was exciting to hear about his lab’s recent use of paired nickase variants of Cas9, which cut just one DNA strand, to induce larger endogenous deletions [13, 14]. Professor Puchta was extremely positive about the potential for GE and in his final perspectives noted that “Synthetic nuclease based DSB-induced DNA repair should be applicable for directed mutagenesis in all transformable plants”, and “in the long run synthetic nuclease-based GE will change plant breeding dramatically”. He also thought it possible that plants with targeted mutations might not be regulated in the same way as transgenic GM plants.
Professor Yang echoed this, presenting a letter from the United States Department of Agriculture (USDA) that informed him that the GE rice produced in his laboratory did not fall within its regulatory authority [14, 15]. Professor Yang documented his work on GE in maize and rice, showing that in cultivars where poor transformation efficiency was a significant bottleneck, GE technologies has sped up the process. He also described the induction of a large deletion of 245 kb in rice using RNA-guided Cas9 [15].
Dr. Laurence Tomlinson and Dr. Vladimir Nekrasov, both from TSL, presented their successful applications of RNA-guided Cas9 nuclease to induce targeted mutagenesis in tomatoes. Tomlinson’s work involved GA signaling whilst Nekrasov described the induction of targeted mutations to engineer pathogen-resistance. He took the audience through initial experimental design, through screening of putatively mutated plants to the identification of individuals showing resistance to powdery mildew. It took just 9 months to identify transgene-free, resistant plants with heritable mutations. Nekrasov confirmed that he and his supervisor, Professor Sophien Kamoun, are now investigating options to make their plants available to growers in regions where the pathogen is a significant problem, whilst also undertaking full-genome sequence analysis to determine if the plants contain any additional mutations. University of Cambridge PhD student, Bernando Pollak, introduced the liverwort Marchantia polymorpha, highlighting the ease by which its genome can be manipulated, as well as its potential as an easily engineerable chassis for synthetic biology. Many of the signaling pathways in Marchantialack the redundancy seen in vascular land plants [16] and so it has huge potential as a tool for the study of plant signaling. Additionally, Marchantia is haploid for a large portion of its life cycle and thus the application of programmable nucleases such as RNA-guided Cas9 are even easier to apply. Dr.Oleg Raitskin (TSL) described experiments to further optimize RNA-guided Cas9 nuclease mediated mutagenesis in plants, including the assessment of orthologues and mutants of Cas9 that may expand the number of possible targets in the genome. He also introduced the concepts behind digital droplet PCR and its implementation in the rapid, quantitative assessment of mutations.
The final presentation was delivered by Edward Perello, Chief Business Officer of Desktop Genetics [17], a UK-based software company who develop tools to support the application of CRISPR-associated technologies. Mr. Perello announced that their guide RNA selection software, Guidebook, now supports six plant genomes (Arabidopsis, rice, maize, wheat, barley and Physcomitrella). Plant scientists were encouraged to use this software, which is free for academics, as well as to contact the Desktop Genetics team with feedback and requests for new features and genomes.
Day two workshop
For the workshop on the second day, participants were given a detailed introduction to the methods used to induce targeted mutagenesis and gene deletions with RNA-guided Cas9 nuclease. This was a hands-on session designed to give the participants a full understanding of how to undertake three key aspects of the technique: selecting target sequences, constructing plasmid vectors, and screening target loci for induced mutations. The content was tailored for researchers working on any transformable plant species.
As well as discussing targeted mutagenesis, Dr. Patron provided an introduction to Type IIS mediated assembly methods for the facile construction of plasmid vectors. Dr. Patron is an advocate for the adoption of standards in bioengineering. She was the lead author on a recent manuscript that described a broadly agreed common genetic syntax for the exchange of DNA parts for plants [18]. In addition, Dr. Patron has contributed to a toolkit of standard parts for plants and created a series of informative online tutorials that introduces users to the Golden Gate Modular Cloning (MoClo) assembly standard [19, 20]. Participants were instructed in the use of published standard parts (Table 1), compatible with the MoClo binary plasmid backbones to build vectors for multiplexed Cas9-induced mutagenesis. The workshop materials have been provided on the GARNet website [21] but the main points are summarized below.
Designing single guide RNAs (sgRNAs) for use with Streptococcus pyogenes Cas9
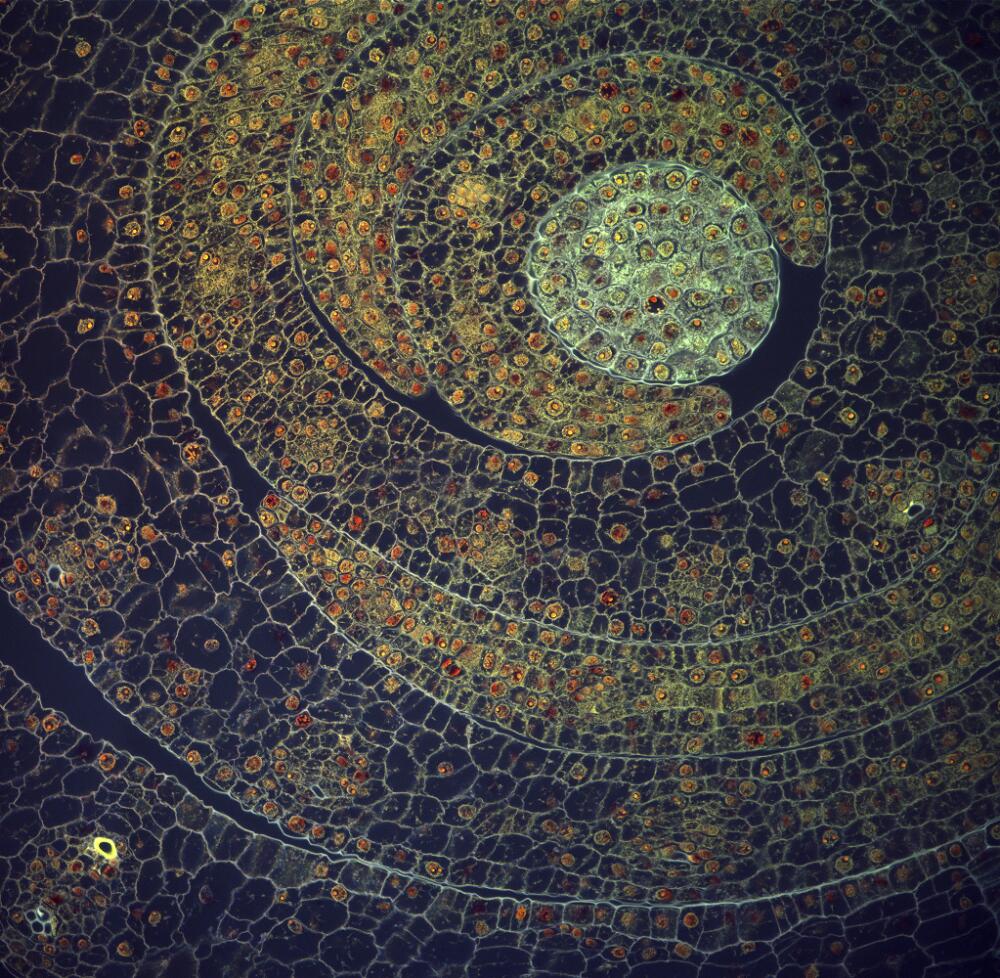 Fernán Federici, OpenPlant Fellow at the University of Cambridge and Director of the Synthetic Biology Lab at Pontificia Universidad Católica in Chile has been awarded a Wellcome Trust Image Award for his micrograph of maize leaves, shot in the Department of Plant Sciences in collaboration with Professor Jim Haseloff.
Fernán Federici, OpenPlant Fellow at the University of Cambridge and Director of the Synthetic Biology Lab at Pontificia Universidad Católica in Chile has been awarded a Wellcome Trust Image Award for his micrograph of maize leaves, shot in the Department of Plant Sciences in collaboration with Professor Jim Haseloff.

![[Closes 24 Nov 2107] Apply now to the OpenPlant Fund!](https://images.squarespace-cdn.com/content/v1/54a6bdb7e4b08424e69c93a1/1509564315902-TUO4I6QRWI9TT8UGSIAJ/OpenPlantTwitter_400x400+%281%29.jpg)

![[Closes 7 Mar 2017] OpenPlant Research Associate (Haseloff Lab)](https://images.squarespace-cdn.com/content/v1/54a6bdb7e4b08424e69c93a1/1486552818859-FH76MCA8SMFU93WB85RX/OpenPlantTwitter_400x400.jpg)








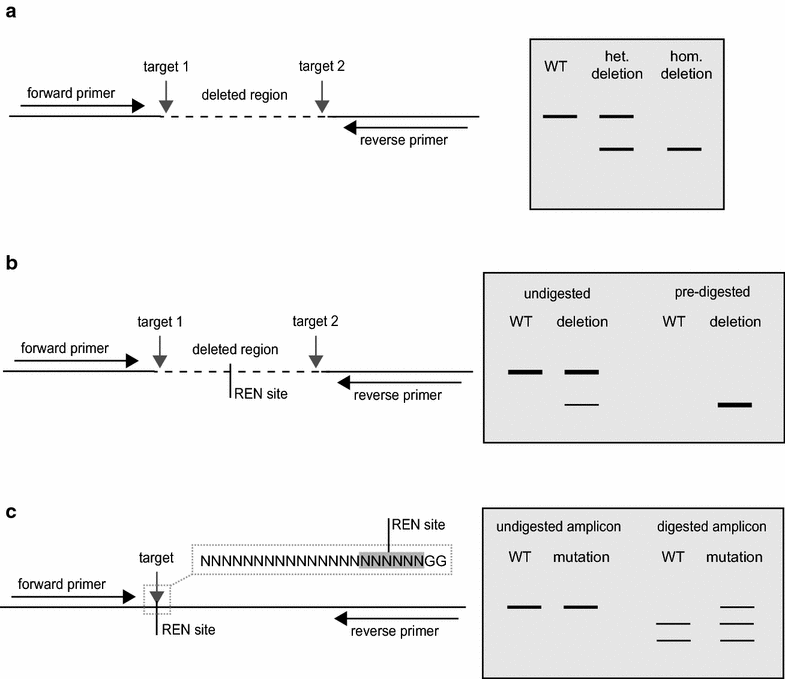
![Fig. 1Interaction of a single guide RNA (sgRNA) expressed from a U6 promoter with its cognate genomic target (adapted from Belhaj et al. [23])](https://images.squarespace-cdn.com/content/v1/54a6bdb7e4b08424e69c93a1/1491867915679-K24J2X8DSXIYA871PON6/image-asset.gif)
 A new
A new 




 The Madagascar Periwinkle (Catharanthus Roseus) - The plant that makes vincristine
The Madagascar Periwinkle (Catharanthus Roseus) - The plant that makes vincristine