The following is a guest post from the SynBio Fund project ‘Development of a microfluidic device for high-throughput analysis of genetic circuits in plant protoplasts’ from by Steven Burgess, Ivan Reyna-Llorens, Christian R. Boehm, Sara Abalde-Cela and Paul Bennett. A continuation project, entitled ‘Establishing 3D Printed Microfluidics for Molecular Biology Workflows’, was funded through the OpenPlant Fund in 2016 and recently a second OpenPlant Fund grant was awarded for the project ‘Plant-ProChip 2.0: High throughput transformation of plant protoplast’.
You can read the original post on their project blog here >>
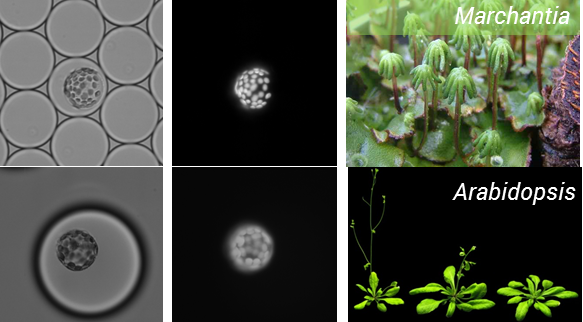
We started this project with the aim of testing whether it is possible to use microfluidics to analyse plant protoplasts, and I think we now have the answer. After numerous rounds of testing we have improved our working method and are now able to routinely isolate and encapsulate protoplasts. This has been done for two model plant species including A. thaliana, and everyone’s favorite Bryophyte –Marchantia polymorphia, the workhorse of the OpenPlant Project for plant synthetic biology (Figure 1).
Figure 1: Encapsulation of protoplasts from model plant species
So the take home message from this project is – if you can protoplast you can encapsulate! But the story does not end here. To be of real use, this process needs to be coupled to transformation of protoplasts. As a result, we teamed up with Oleg Raitskinfrom Nicola Patron’s group at the Earlham Institute. Oleg has been optimizing protoplast isolation and transformation using Nicotiana benthamiana and had a couple of tips for improving isolation, including the use of a cork borer instead of scalpel blade for cutting up tissues to minimize mechanical damage, cutting tissue when submerged in the enzyme mix and using a high ratio of DNA to protoplasts during PEG transformation.
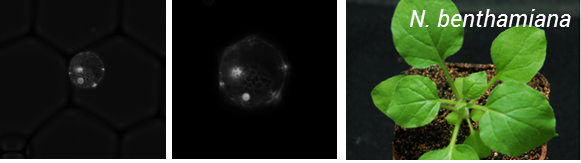
This was a fruitful collaboration, Oleg managed to transform protoplasts with a nuclear targeted Venus reporter and these were encapsulated by Ziyi in the Chemistry department (Figure 2).
Figure 2: Encapsulation of N. benthamiana protoplats expressing a nuclear targeted Venus reporter
So putting all this work together, we have in hand a simple, but very powerful system that opens up a whole range of possibilities for rapid phenotyping in plants (Figure 3).
Figure 3: Schematic of microfluidic analysis of plant protoplasts and some of the potential applications.
One of the stipulations of the project was to pursue science in an open manner, so we have been putting up information on the website protocols.io. I highly recommend checking out the site if you haven’t done so already, it has a great set up for disseminating protocols. Further we believe microfluidics is a great technique, so would encourage others to have a go as well!
Looking to the future there are still a few things we would like to work on, The project was briefly presented at Cambridge’s Cafe Synthetique meet-up and we had some great feedback, such as trying Calcium alginate encapsulation as a means of improving protoplast viability. Sorting of protoplasts is the next major goal, and requires redesign of a new chip, and finally improving the efficiency of protoplast transformation by developing an on-chip procedure would be a big advantage. This round of our project has come to an end, but stay tuned for future developments.
Finally I want to finish this piece with a big thanks to Cambridge Synthetic Biology SRI for funding the work, it has been a great experience, and to encourage anyone who is interested in protoplasts or microfluidics to get in contact, we are always happy to chat!

![[Closes 24 Nov 2107] Apply now to the OpenPlant Fund!](https://images.squarespace-cdn.com/content/v1/54a6bdb7e4b08424e69c93a1/1509564315902-TUO4I6QRWI9TT8UGSIAJ/OpenPlantTwitter_400x400+%281%29.jpg)

![[Closes 7 Mar 2017] OpenPlant Research Associate (Haseloff Lab)](https://images.squarespace-cdn.com/content/v1/54a6bdb7e4b08424e69c93a1/1486552818859-FH76MCA8SMFU93WB85RX/OpenPlantTwitter_400x400.jpg)