Digital tools
Workpackage E: Digital tools for plant synthetic biology
Software tools play an increasingly important role in Synthetic Biology experiments, as we automate experiments, and as the systems we construct increase in scale. In order to accurately predict the behaviour of biological systems, which are governed by multi-scale, parallel, and feedback-regulated interactions, we need computational models. OpenPlant aims to provide software to (i) automate DNA assembly, (ii) quantify gene expression in plants, and (iii) create models for gene expression and cell growth.
Air chambers on the top surface of the Marchantia polymorpha thallus
Image: Jim Haseloff
The Marchantia Genome Database: MarpoDB
In addition to its role as a gene-centric database for finding plant DNA parts, the Marchantia genome database MarpoDB (Delmans et al., 2017) contains gene models with predicted transcripts, encoded proteins and phylogenetic comparison data. It maintains links to the Tak1 Marchantia reference genome and nomenclature, and allows interpretation of transcriptomic data. It has also formed the basis for analysis of differential gene expression in germinating Marchantia spores.
Future versions of the database will incorporate more features for describing characterisation of parts and gene expression using Plant Ontology terms. The online database can be found at: http://www.marpodb.io
Green and red fluorescent protein expressing E. coli cells in a biofilm
Image: Fernan Federici
Quantitative image analysis and model building
The combination of imaging data and high-speed computing allows the generation of software models for complex genetic networks and physico-genetically coupled multicellular systems. The behaviour of these systems cannot be predicted easily, and software models are essential tools for both understanding their properties, and for building new networks. In particular, DNA-based reprogramming of cellular networks would allow rational design of new crops, feedstock and cellular therapies.
CellModeller is a Python-based framework for modelling large-scale multi-cellular systems, such as biofilms, plant and animal tissue. Members of the Haseloff Lab have developed models of cellular biophysics, gene regulation and other intracellular processes, and intercellular signalling. The idea of CellModeller is to create a system to simulate these models together in populations of growing and dividing cells. The latest features include cellcell adhesion and cell shape, as well as algorithms for whole colonyscale segmentation from confocal microscopy datasets.
CellModeller is open source and on Github. There is a support googlegroup.
Dynamic software models and gene expression
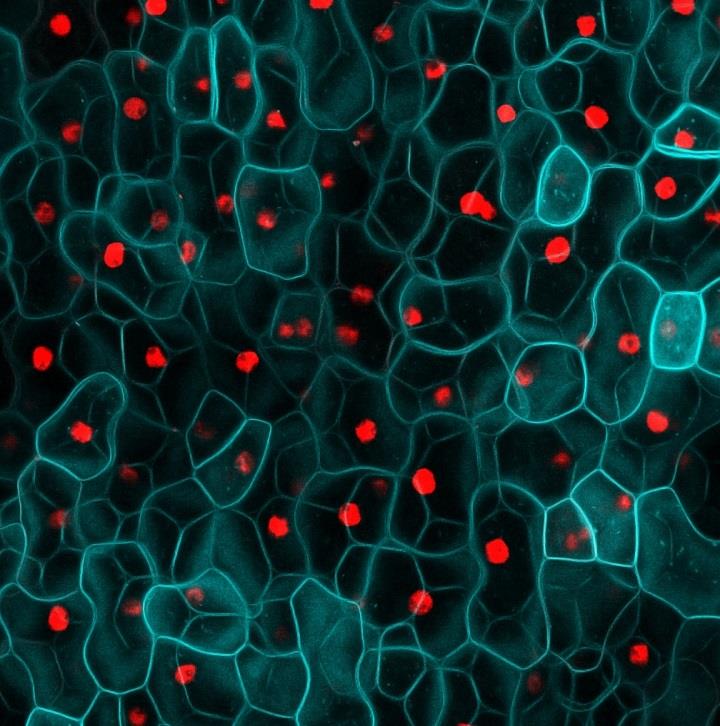
In planta cytometry in Arabidopsis thaliana
Image: Fernan Federici
The combination of specific labeling with gene markers and advanced microscopy allows extraction of quantitative information from intact biological samples. Gene expression can be integrated with cellular geometry, combined with other markers and mapped over time. This can produce parameter sets that directly inform executable models of biological systems. This a fruitful area for collaboration between biologists and physicists and mathematicians.
The Haseloff lab has developed three-parameter measurement techniques for quantifying gene expression in cell suspensions in such as way that extrinsic noise is minimised and a reliable estimate of the intrinsic properties of gene promoters can be made (Rudge et al., 2016; Grant et al., 2016). This relies on software models for gene expression, cell growth, and the use of a co-expressed marker to reduce variation. A computational framework has been established to allow automated analysis of microplate reader data, and this has been made available on Github.
Martins et al. (2018) Fig 1: Multiple internal and external factors coordinate cell growth and divisions in cyanobacteria.
Synthetic circuits and modelling to rewire the circadian clock
The Locke lab has used synthetic circuits to rewire the cyanobacterial circadian clock. They used constructs to probe the circadian clock and its outputs at the single cell level in S. elongatus, and built models of the coupling of the cyanobacterial clock, that are publicly available at the Molecular Systems Biology website and are written in SBML to enable easy sharing of the code. The Locke lab used iterative cycles between stochastic modelling and experiments to understand how the circadian clock and environmental cycles modulate cell size control and division timings. Read more about their work in Martins et al. (2018).
Simarly, the Locke lab has used a combination of modelling and experimentation to better understand the circadian clock in Arabidopsis thaliana. Read more about their recent work on this in Greenwood et al., (2019).
Publications
Ho PY, Martins BMC, Amir A. (2020) A Mechanistic Model of the Regulation of Division Timing by the Circadian Clock in Cyanobacteria. Biophysical Journal. Volume 118, Issue 12, Pages 2905-2913, https://doi.org/10.1016/j.bpj.2020.04.038.
Grant PK, Szep G, Patange O, Halatek J, Coppard V, Csikász-Nagy A, Haseloff J, Locke JCW, Dalchau N & Phillips A. (2020). Interpretation of morphogen gradients by a synthetic bistable circuit. Nature Communications 11: 5545 https://doi.org/10.1038/s41467-020-19098-w
Boehm CR, Grant PK, Haseloff J. (2018). Programmed hierarchical patterning of bacterial populations. Nat Commun. 9(1):776. doi: 10.1038/s41467-018-03069-3.
Delmans M, Haseloff J, (2018). µCube: A Framework for 3D Printable Optomechanics. Journal of Open Hardware, 2. 10.5334/joh.8.
Kan A, Del Valle I, Rudge T, Federici F, Haseloff J. (2018). Intercellular adhesion promotes clonal mixing in growing bacterial populations. J R Soc Interface. 15(146). pii: 20180406. doi: 10.1098/rsif.2018.0406.
Martins BMC, Tooke AK, Thomas P, Locke JCW, (2018). Cell size control driven by the circadian clock and environment in cyanobacteria. Proc Natl Acad Sci U S A. 115(48):E11415-E11424. doi: 10.1073/pnas.1811309115.
Delmans M, Pollak B, and Haseloff J., (2017). MarpoDB: An open registry for Marchantia polymorpha genetic parts. Plant Cell Physiol. 58(1):e5. doi: 10.1093/pcp/pcw201.
Nuñez IN, Matute TF, Del Valle ID, Kan A, Choksi A, Endy D, Haseloff J, Rudge TJ, Federici F (2017). Artificial Symmetry-Breaking for Morphogenetic Engineering Bacterial Colonies. ACS Synth Biol. 6(2):256-265. doi: 10.1021/acssynbio.6b00149.
Grant PK, Dalchau N, Brown JR, Federici F, Rudge TJ, Yordanov B, Patange O, Phillips A, Haseloff J (2016). Orthogonal intercellular signaling for programmed spatial behavior. Mol Syst Biol. 12(1):849. doi: 10.15252/msb.20156590.
Martins BM, Das AK, Antunes L, Locke JC (2016). Frequency doubling in the cyanobacterial circadian clock. Mol Syst Biol. 12(12):896. doi: 10.15252/msb.20167087.
Rudge T, Brown J, Federici F, Dalchau N, Phillips A, Ajioka J, & Haseloff J (2016). Characterization of intrinsic properties of promoters. ACS Synth Biol. 5(1):89-98. doi: 10.1021/acssynbio.5b00116.